Genomic stability can be maintained through Homologous Recombination Repair (HRR) when DNA double-strand breaks occur in normal cells, but mutations in HRR-related genes can cause Homologous Recombination Deficiency (HRD), which manifests as Genomic Scar (GS).
The detection of HRD can be used to assess the risk of cancers, including breast, ovarian, and pancreatic cancers, etc. Also, since most HRD tumors are highly sensitive to PARP inhibitors (PARPi) and platinum drugs, the results of HRD detection can be used to predict the efficacy and provide the clinical medication guidance.
Traditional strategy of differential capture
A comprehensive HRD assessment consists of variant detection of HRR-related genes and genomic scar identification based on SNPs loci. From the perspective of clinical testing accuracy, HRR gene detection and genomic scar identification based on SNPs loci generally require no less than 500x and 100x effective sequencing depths respectively, to detect mutations with a frequency of no less than 1% and SNPs loci with high MAF values in healthy Chinese populations.
For differential capture, the traditional strategy is to control the ratio of different probes during hybridization, but the molar concentration of probes relative to the template molecules is excessive, making it time-consuming and laborious to adjust the probe concentration ratio to achieve differential capture, and the results are unstable and do not achieve good isobaric scaling.
The Dark Probe technology makes differential capture visible and easy
Compared to traditional strategy, Dynegene's self-developed Dark Probe technology makes complex differential capture easier. The Dark Probe technology not only shortens the optimization period and saves cost and labor, but also optimizes the instability of results caused by the traditional strategy of controlling probe ratio, and has high intra-batch and inter-batch consistency.
More importantly, the high precision result can be easily scaled according to the customers’ various needs of differential capture, which is a promising application.
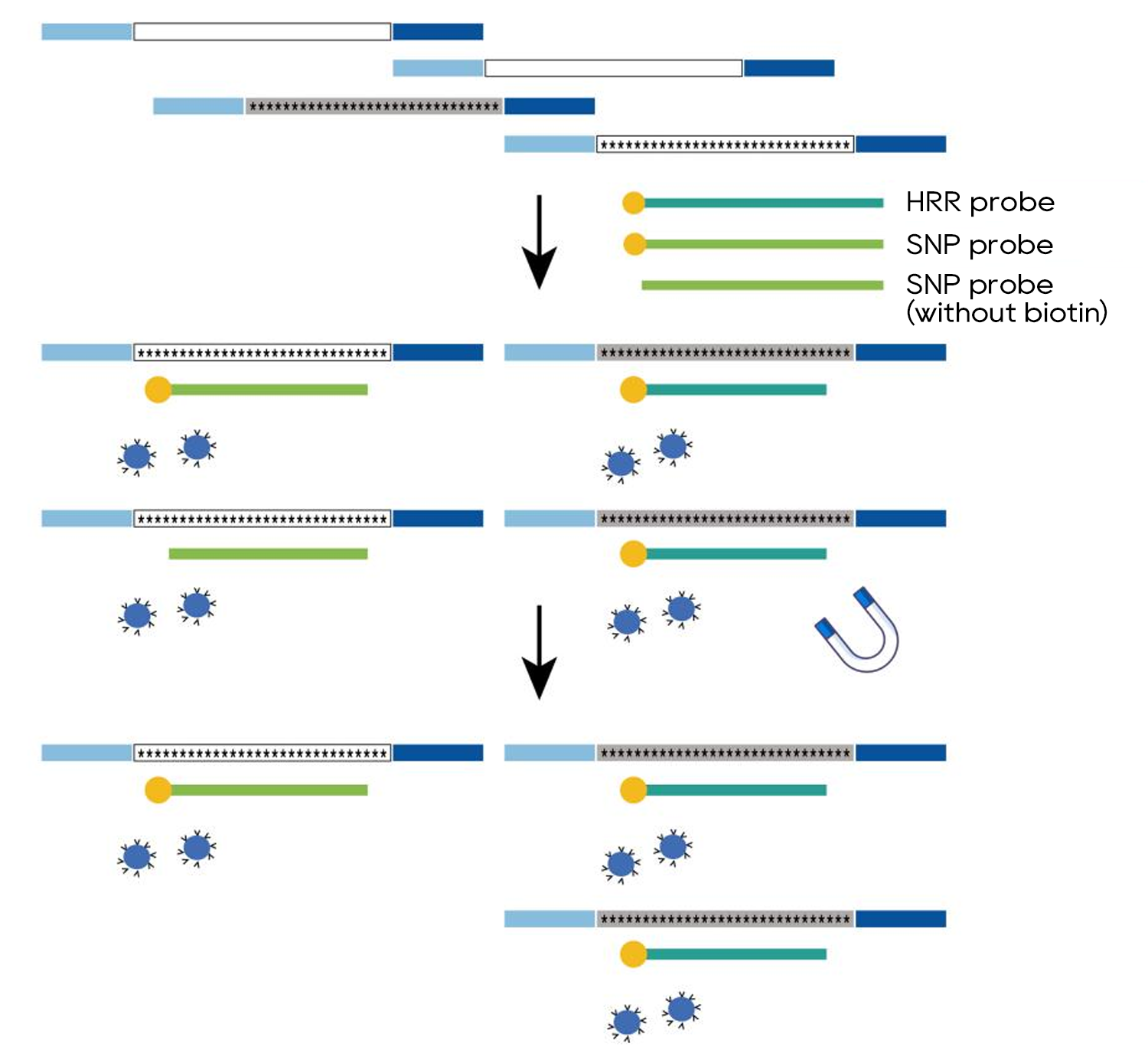
Workflow of Dark Probe
Product Highlights
Comprehensive testing
20 HRR-related core genes are selected based on NCCN guidelines and clinical trial experience; 50,000 SNPs with high MAF values are also collected to cover as many heterozygous loci as possible for genomic scar identification.
Shorter optimization period for custom needs
For differential capture needs, the Dark Probe technology significantly shortens the optimization period, and can easily achieve isobaric scaling according to various differential capture needs of customers.
Excellent Consistency
The Dark Probe technology optimizes the instability of results caused by the traditional strategy of controlling probe ratios, and provides excellent consistency for intra-batch and inter-batch.
Lower Cost
Differential capture directly leads to a significant reduction in sequencing costs.
Data demonstration
Good performance of capture
Input 100 ng of standard (NA12878) for ultrasonic shearing and DNA library construction (QuarPrep DNA Library Kit, Dynegene, NL1001); 750 ng of constructed library treated by conventional hybridization capture using QuarXeq HRD probe (QuarHyb One Reagent Kit, Dynegene, NC1003), three replicate experiments, sequencing volumn of 6G on illumina platform.
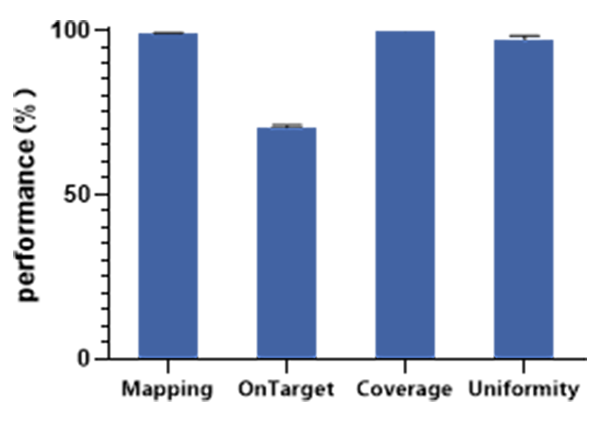
Figure 1. Capture Performance of QuarXeq HRD Probes
The ratio of HRR and HRD depth under differential capture is consistent with the expectation
Input 100 ng of standard (NA12878) for ultrasonic shearing and DNA library construction (QuarPrep DNA Library Kit, Dynegene, NL1001); 750 ng of constructed library treated by conventional hybridization capture using QuarXeq HRD Probes probe (QuarHyb One Reagent Kit, Dynegene, NC1003), three replicate experiments, illumina platform sequencing. The analysis results showed that the ratio of HRR depth to HRD depth was 5:1, as expected.
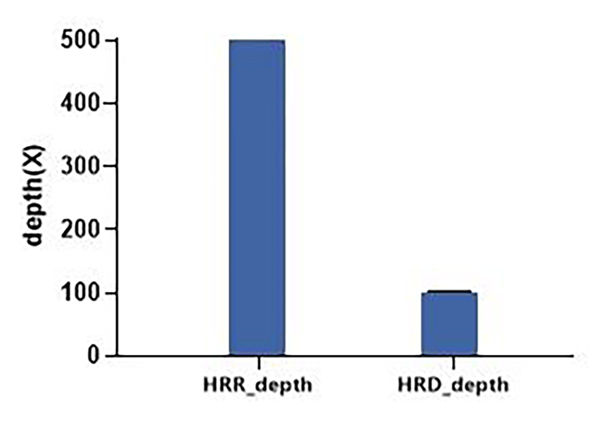
Figure 2. Depth under Differential Capture Using QuarXeq HRD Probes
The HRD BRCA2 depth and Chr 13 depth were also analyzed for the BRCA2 gene, and the results showed a ratio of 5:1, also as expected.
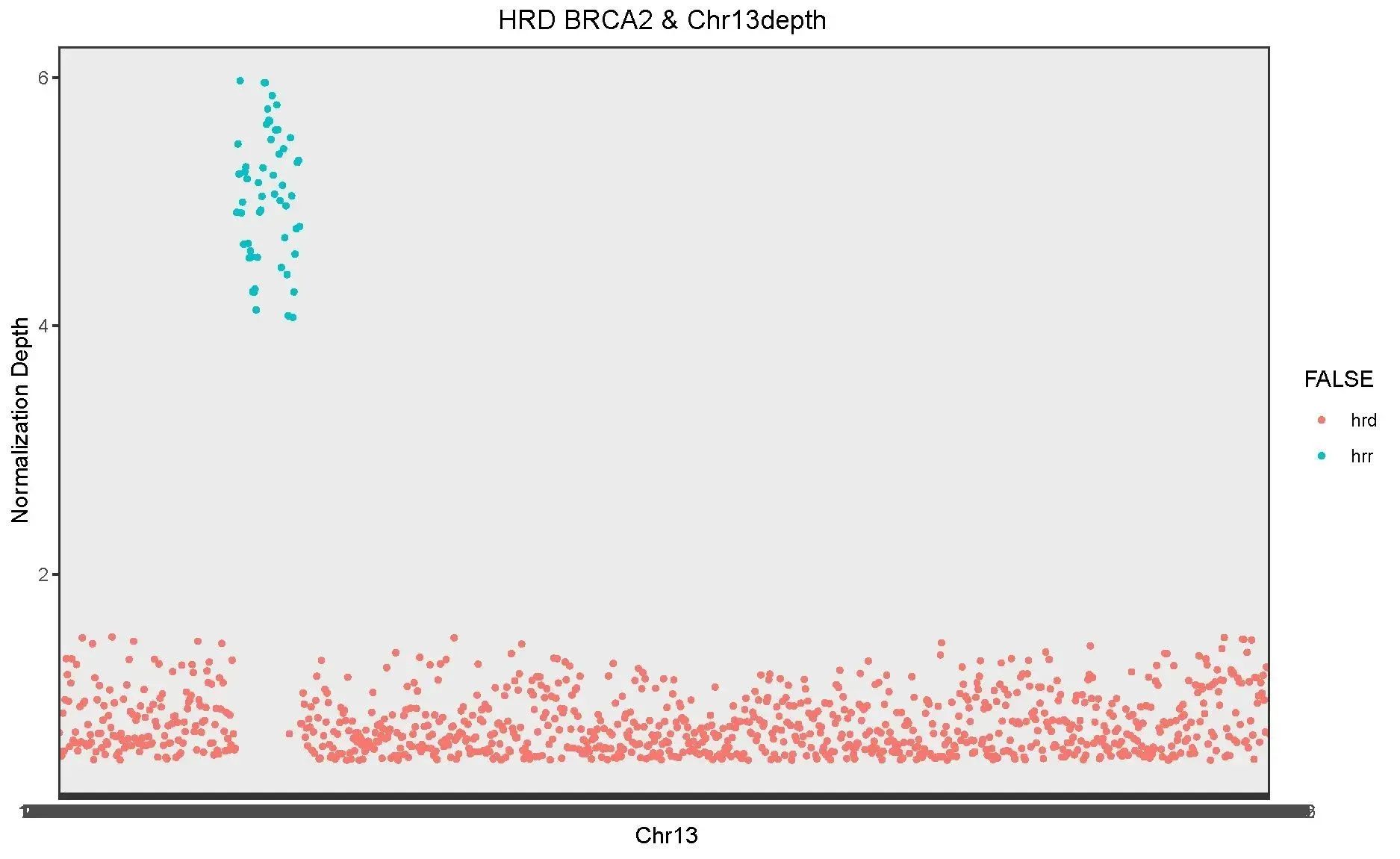
Figure 3. Comparison of HRD BRCA2 depth and CHR 13 depth under differential capture using QuarXeq HRD Probes
Uniform coverage of SNPs loci
The SNPs loci in QuarXeq HRD Probes are evenly distributed to cover as many heterozygous loci as possible, ensuring that LOH, LST, TAI that meet the requirements can be detected.
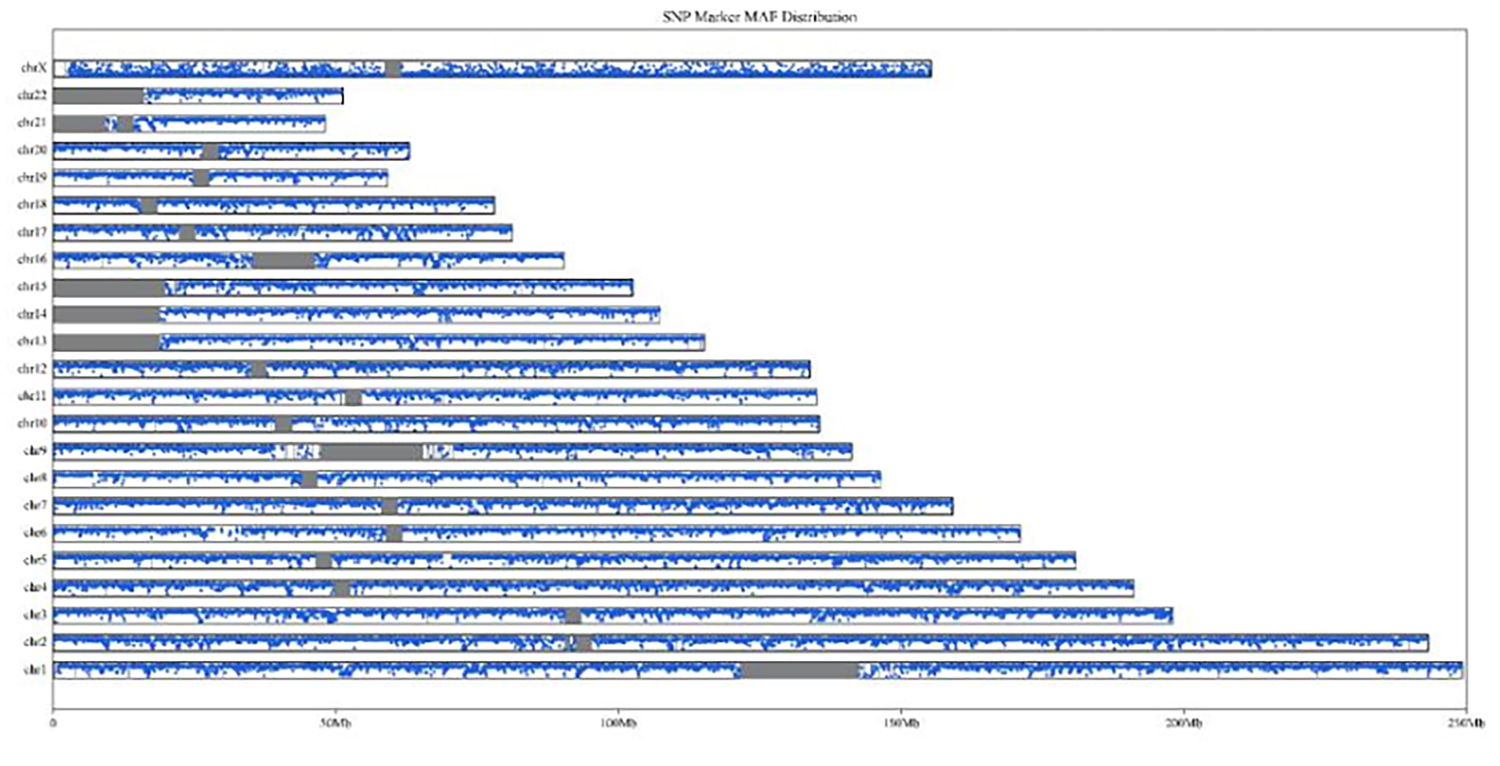
Figure 4. Coverage of SNP Marker chromosomes
Note: the gray area is the highly repetitive region on the reference genome, and the blue scatter is the SNP Marker MAF repeat element.
HRR-related genes
Ordering information

 NGSHybridization Capture DNA Probe QuarStar Liquid Pan-Cancer Panel 3.0 QuarStar Pan-Cancer Lite Panel 3.0 QuarStar Pan-Cancer Fusion Panel 1.0 QuarStar Pan Cancer Panel 1.0 Hybridization Capture RNA Probe QuarXeq Human All Exon Probes 3.0 HRD panel Library Preparation DNA Library Preparation Kit Fragmentation Reagent mRNA Capture Kit rRNA Depletion Kit QuarPro Superfast T4 DNA Ligase Hybridization Capture QuarHyb Super DNA Reagent Kit QuarHyb DNA Plus 2 Reagent Kit QuarHyb DNA Reagent Kit Plus QuarHyb One Reagent Kit QuarHyb Super Reagent Kit Pro Dynegene Adapter Family Dynegene Blocker Family Multiplex PCR QuarMultiple BRCA Amplicon QuarMultiple PCR Capture Kit 2.0 PathoSeq 450 Pathogen Library Corollary Reagent Streptavidin magnetic beads Equipment and Software The iQuars50 NGS Prep System
NGSHybridization Capture DNA Probe QuarStar Liquid Pan-Cancer Panel 3.0 QuarStar Pan-Cancer Lite Panel 3.0 QuarStar Pan-Cancer Fusion Panel 1.0 QuarStar Pan Cancer Panel 1.0 Hybridization Capture RNA Probe QuarXeq Human All Exon Probes 3.0 HRD panel Library Preparation DNA Library Preparation Kit Fragmentation Reagent mRNA Capture Kit rRNA Depletion Kit QuarPro Superfast T4 DNA Ligase Hybridization Capture QuarHyb Super DNA Reagent Kit QuarHyb DNA Plus 2 Reagent Kit QuarHyb DNA Reagent Kit Plus QuarHyb One Reagent Kit QuarHyb Super Reagent Kit Pro Dynegene Adapter Family Dynegene Blocker Family Multiplex PCR QuarMultiple BRCA Amplicon QuarMultiple PCR Capture Kit 2.0 PathoSeq 450 Pathogen Library Corollary Reagent Streptavidin magnetic beads Equipment and Software The iQuars50 NGS Prep System Primers and Probes
Primers and Probes RNA SynthesissgRNA miRNA siRNA
RNA SynthesissgRNA miRNA siRNA



 Gene Synthesis
Gene Synthesis Oligo Pools
Oligo Pools CRISPR sgRNA Library
CRISPR sgRNA Library Antibody Library
Antibody Library Variant Library
Variant Library









 Tel: 400-017-9077
Tel: 400-017-9077 Address: Floor 2, Building 5, No. 248 Guanghua Road, Minhang District, Shanghai
Address: Floor 2, Building 5, No. 248 Guanghua Road, Minhang District, Shanghai Email:
Email: Tel: 400-017-9077
Tel: 400-017-9077 Address: Floor 2, Building 5, No. 248 Guanghua Road, Minhang District, Shanghai
Address: Floor 2, Building 5, No. 248 Guanghua Road, Minhang District, Shanghai Email:
Email: 







